Software
DeepChopper

DeepChopper: Genomic language model to identify chimera artifacts in long-read direct RNA sequencing data
OctopusV

OctopuSV: Advanced structural variant analysis toolkit
PxBLAT
An Efficient and Ergonomic Python Binding Library for BLAT
ScanExitronLR
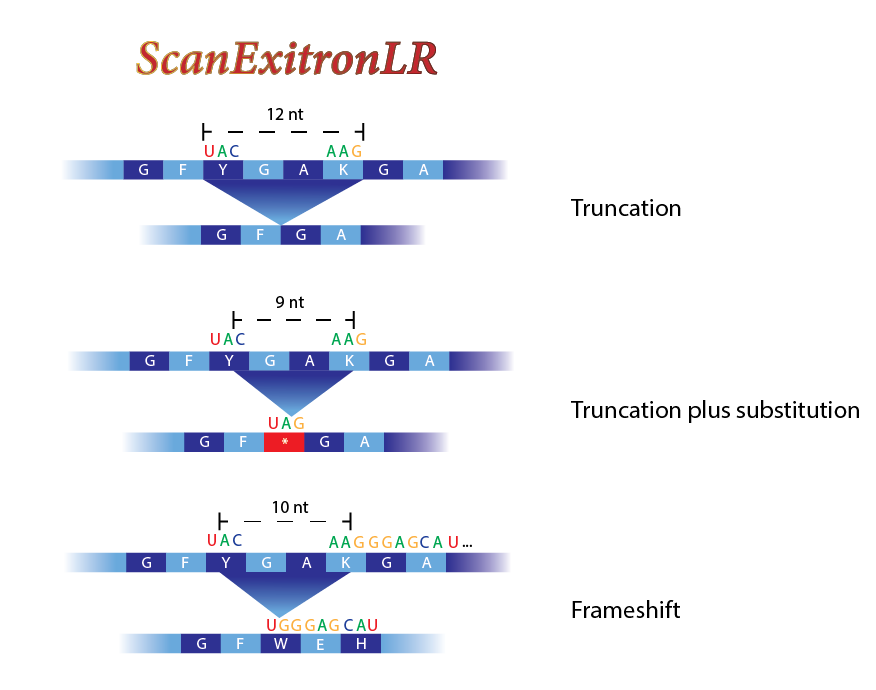
LncGSEA is a computational workflow for exitron splicing identification in long-read RNA-seq data.
LncGSEA

LncGSEA is a convenient tool to predict the lncRNA associated pathways through Gene Set Enrichment Analysis (GSEA) of gene expression profiles from large-scale cancer patient samples.
ScanExitron
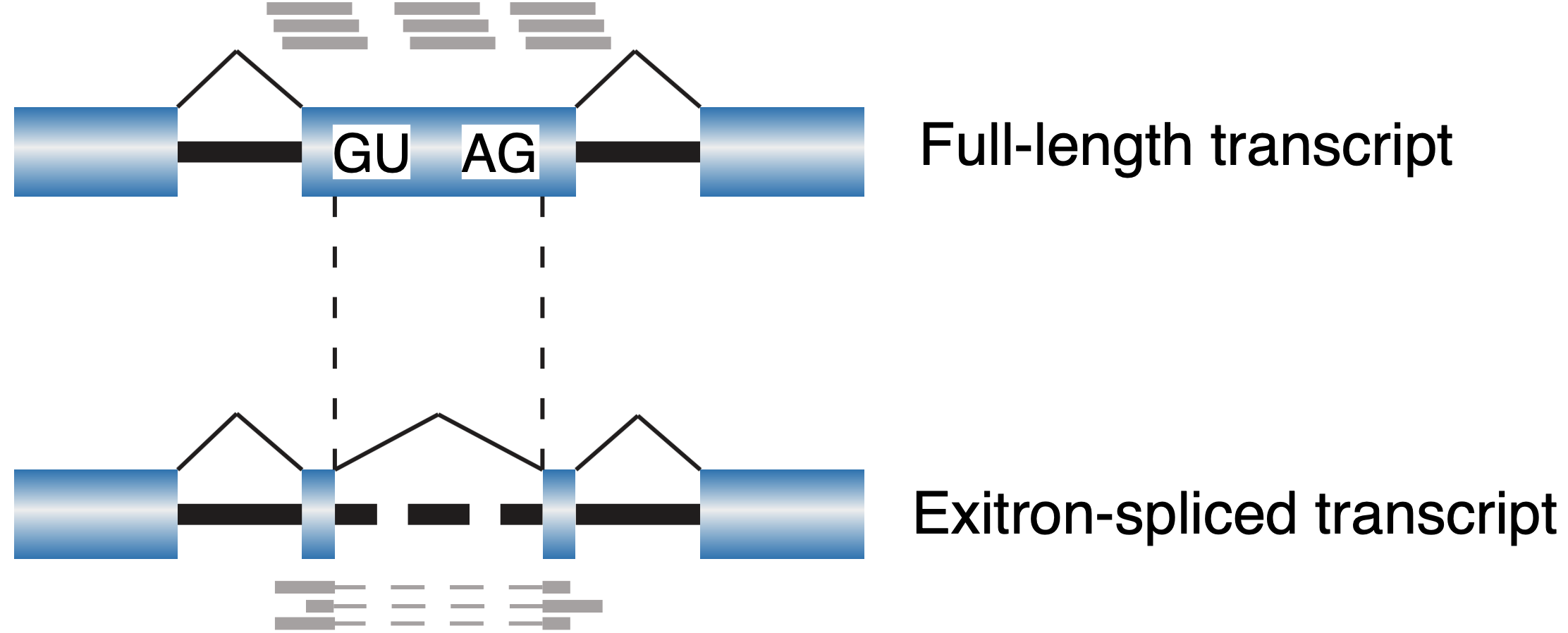
ScanExitron is a computational workflow for exitron splicing identification from RNA-seq.
ScanITD

ScanITD performs a stepwise seed-and-realignment procedure for internal tandem duplication (ITD) detection with accurate variant allele fraction prediction.
ScanNeo
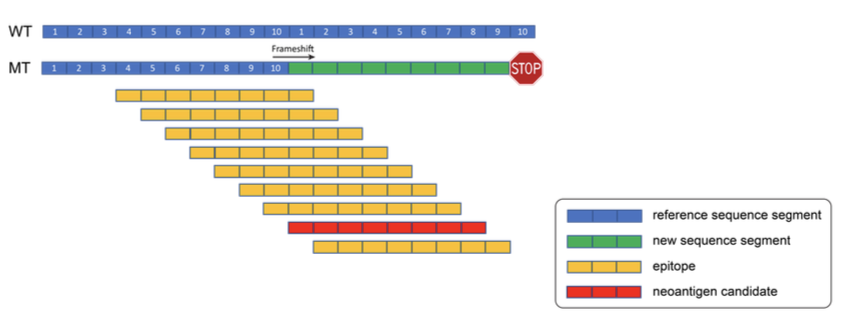
ScanNeo is a pipeline for identifying insertion and deletion (indel) introduced neoantigens from RNA sequencing data.
transIndel
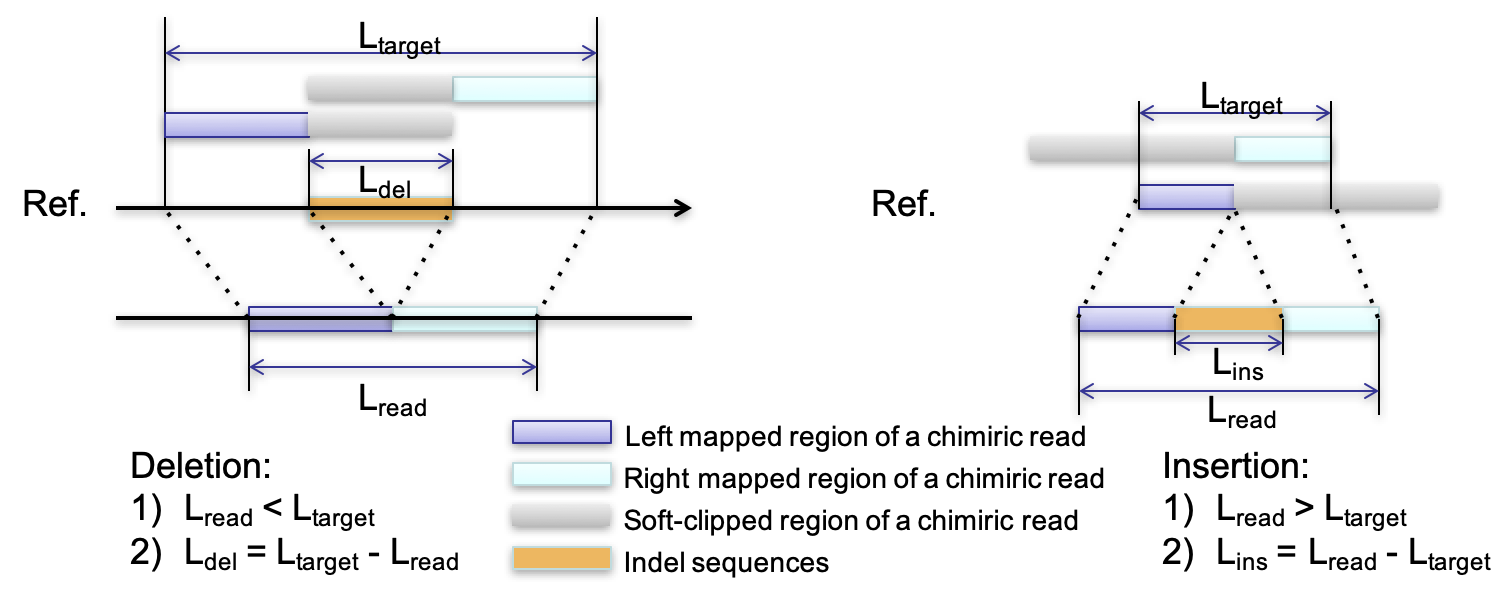
transIndel is used to detect indels (insertions and deletions) from DNA-seq or RNA-seq data by parsing chimiric alignments from BWA-MEM.
ScanIndel
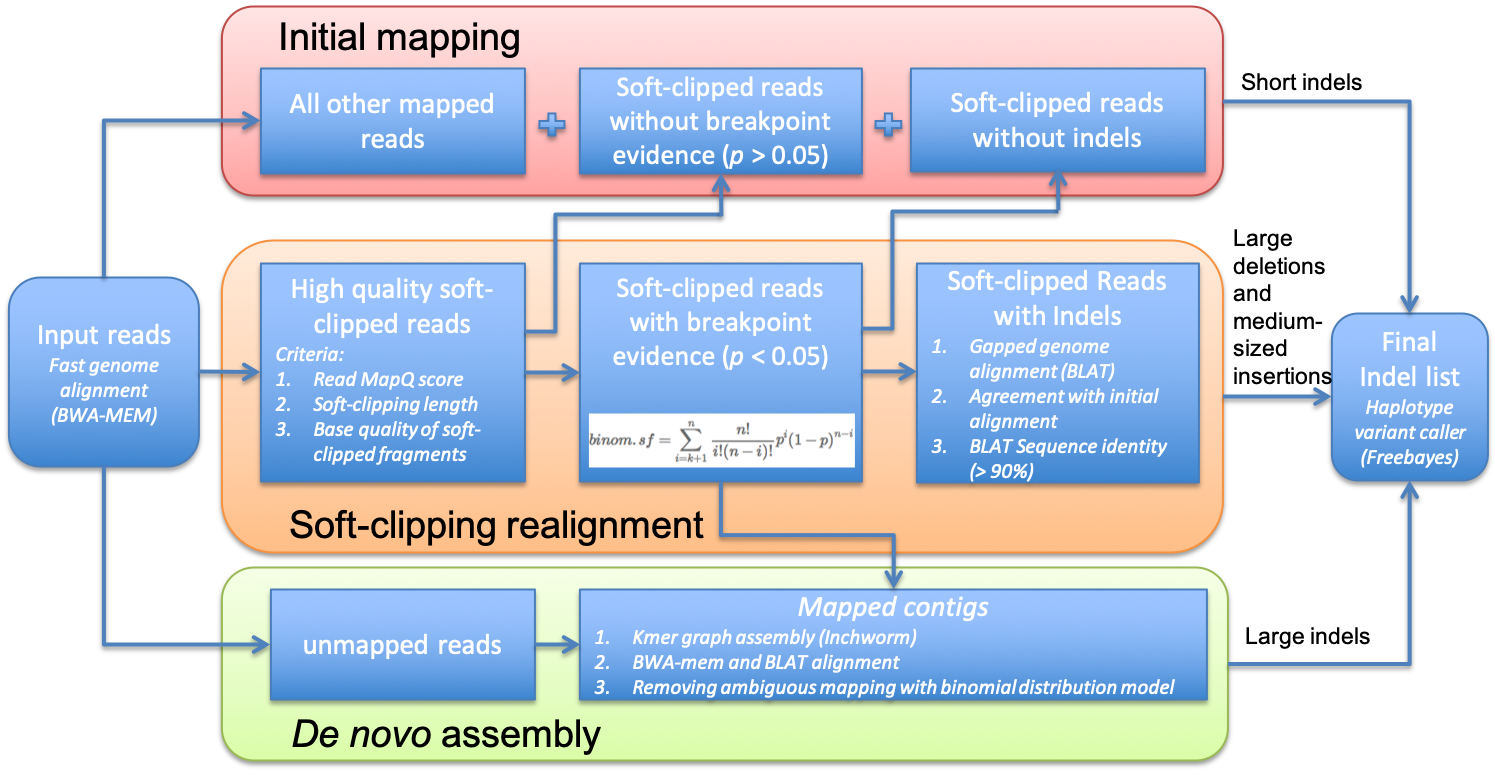
ScanIndel is a python program to detect indels (insertions and deletions) from NGS data by re-align and de novo assemble soft clipped reads.
EgoNet
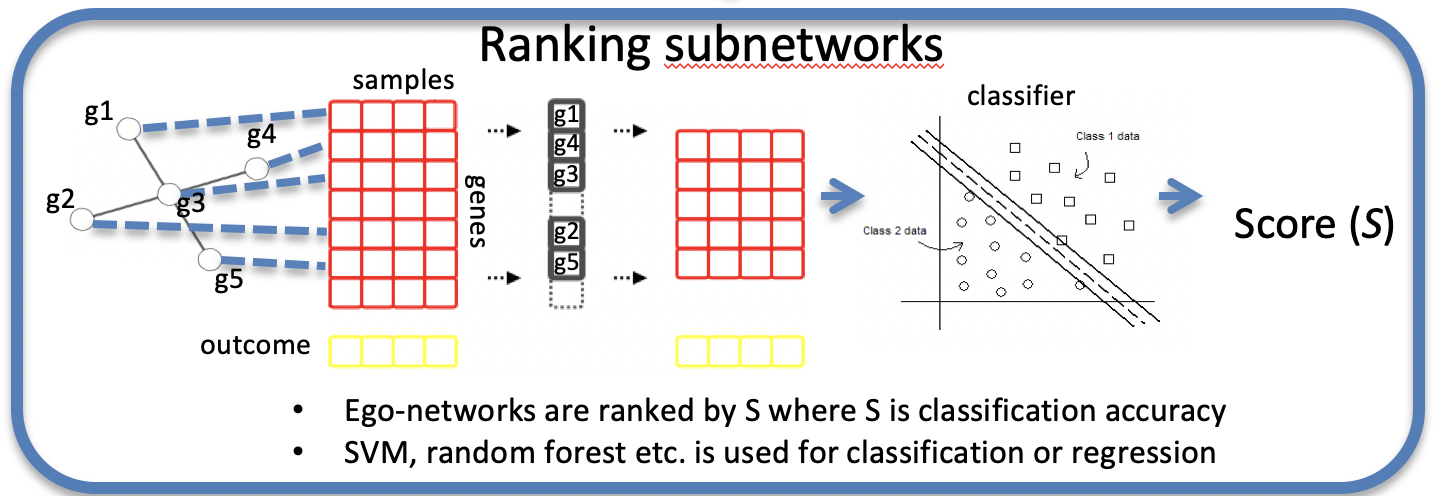
EgoNet is implemented by Python and it is designed to detecting disease related subnetwork from a large biological network (PPI, metabolic network) combined with gene expression data.
ARSER
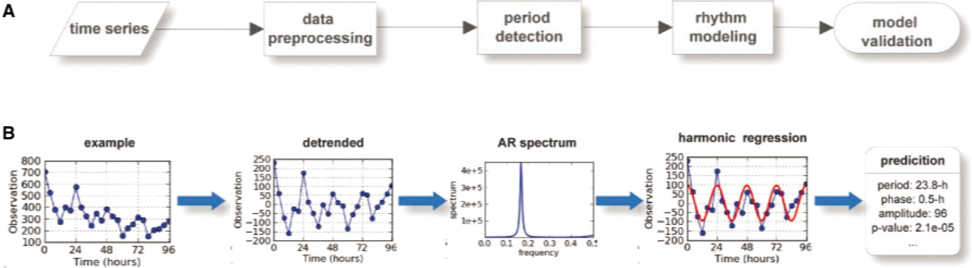
ARSER is a Python package for identifying periodic expression profiles in analyzing circadian microarray data and has been released under the GPL.