Research Highlights
Genomic alterations detection
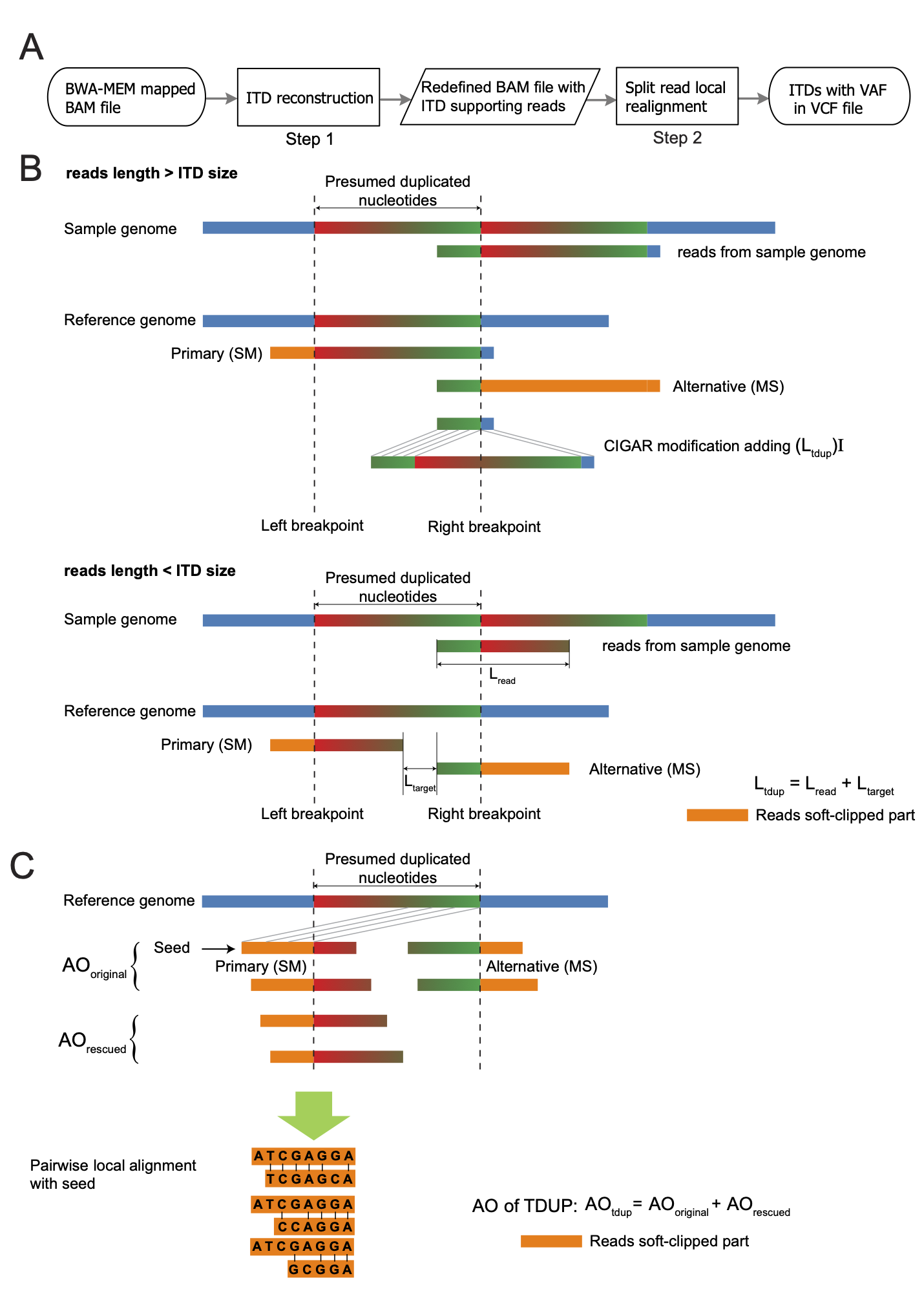
We have developed multiple algorithms specilized in analyzing high-throughput sequencing data to detect DNA alterations including insertions and deletions (INDELs) and large-scale structural variations (SVs). We developed the ScanIndel algorithm to address the challenge of reliably detecting medium- and large-sized indels from whole-exome or whole-genome sequencing data. Our contributions for SV detection are highlighted by two algorithms developed in my group: SVfinder and ScanITD. Application of these algorithms in cancer genomic data, we were able to detect diverse genomic rearrangement events involving androgen receptor (AR) in prostate cancer as well as a t(11;17) translocation event causing the SPI-ZNF287 gene fusion in multiple myeloma.
Reference
- Wang TY, Yang R. ScanITD: Detecting internal tandem duplication with robust variant allele frequency estimation, GigaScience, Volume 9, Issue 8, August 2020, giaa089.
- Yang R, Nelson AC, Henzler C, Thyagarajan B, Silverstein KA. ScanIndel: a hybrid framework for indel detection via gapped alignment, split reads and de novo assembly. Genome Medicine. 2015 Dec;7(1):1-2.
Transcriptomic mis-splicing events discovery
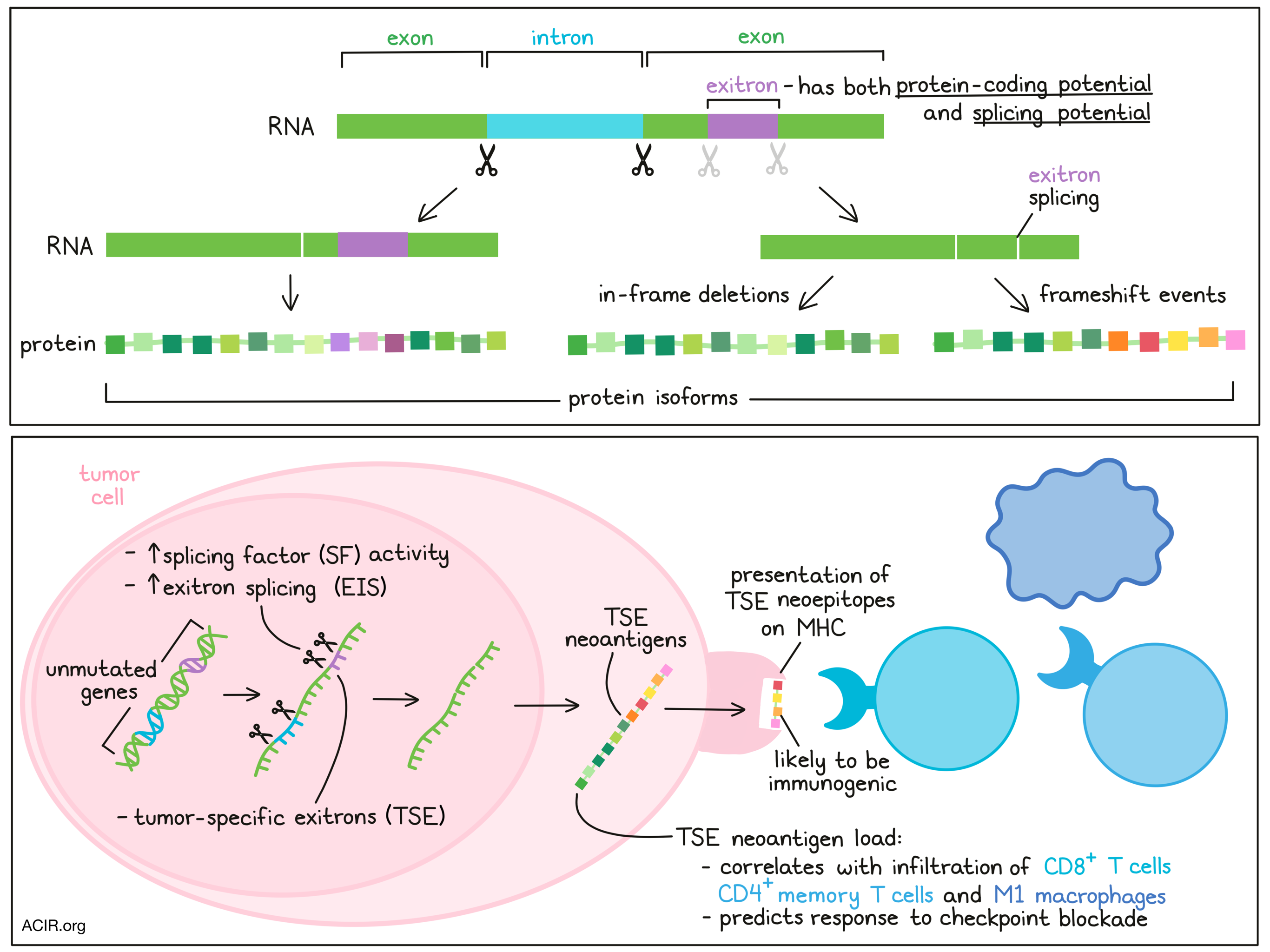
We were the first to discover a type of non-canonical splicing named “exitron” that result in internally deleted protein sequences from annotated coding exons in prostate cancer. Through integrated Pan-Cancer analysis, we observed exitron splicing contributes oncogenic phenotype and represents a source for a new set of neoantigens that are potentially targetable with immunotherapy. Moreover, we identified various types of RNA mis-splicing and alterations, such as cryptic exon, alternative polyadenylation and non-linear splicing in AR that contributed constitutive activity of the broad AR transcriptional program. These activities resulted from AR splicing variants are completely insensitive to all current prostate cancer targeted therapies, including the second-generation AR antagonist enzalutamide.
Multi-omics integrative analysis
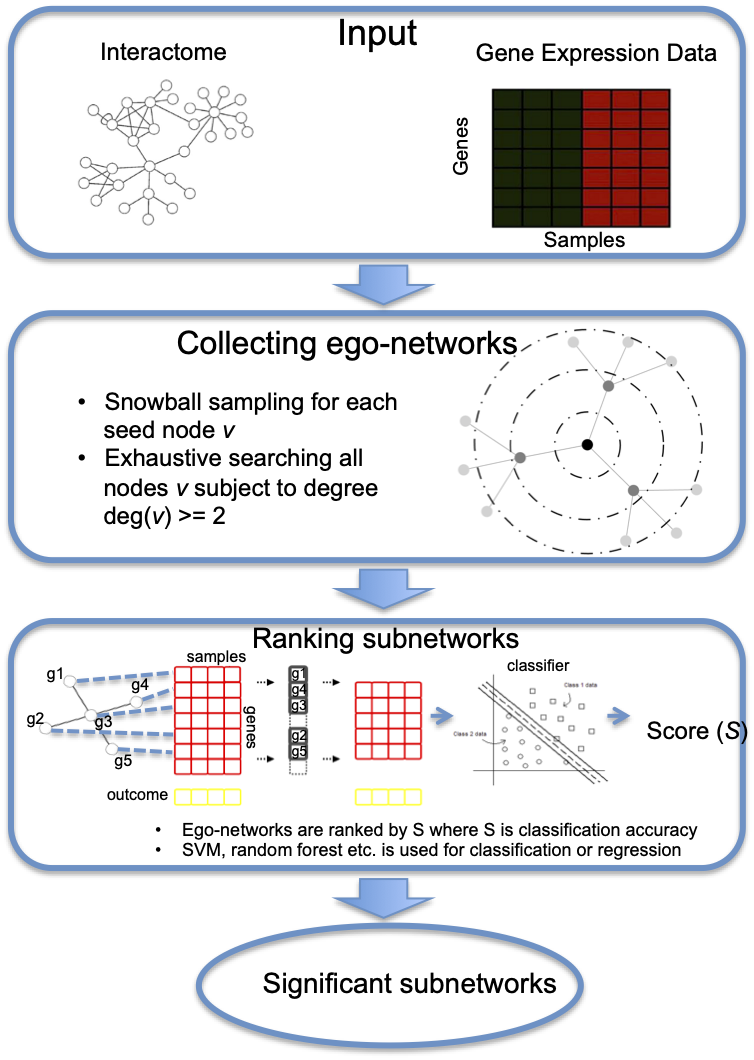
A major goal of our research is to develop integrative computational tools that leverage multi-omics biological and clinical datasets to infer disease-associated genes, regulatory interactions and patient-specific therapeutic targets. Our contributions in this area include: 1) developing the EgoNet algorithm by integrating gene expression data and protein-protein interaction networks to identify gene markers that are associated with clinical phenotypes; 2) Constructing transcriptional regulatory networks of major transcriptional factors AR, BRD4 and STAT5 with ChIP-seq and gene expression data that drive prostate cancer and leukemia and 3) Integrating transcriptomic and proteomic data to predict patient-specific tumor neoantigens for immunotherapy treatment.
Reference
- Wang TY, Wang L, Alam SK, Hoeppner LH, Yang R. ScanNeo: identifying indel derived neoantigens using RNA-Seq data. Bioinformatics. 2019 Mar 18.
- Yang R, Bai Y, Qin Z, Yu T. EgoNet: identification of human disease ego-network modules. BMC Genomics. 2014 Apr 28;15:314.